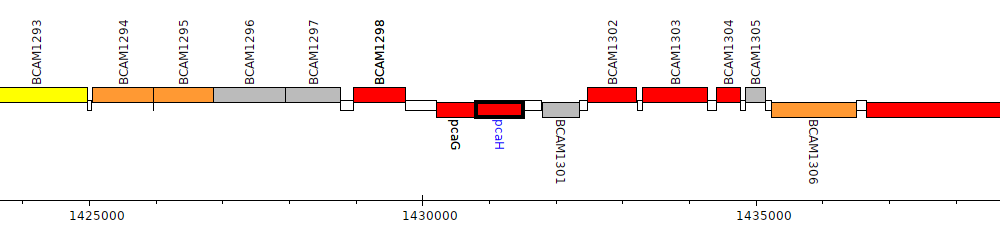
Burkholderia cenocepacia J2315, BCAM1300 (pcaH)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0019619 | 3,4-dihydroxybenzoate catabolic process |
Inferred from Sequence Model
Term mapped from: InterPro:cd03464
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0018578 | protocatechuate 3,4-dioxygenase activity |
Inferred from Sequence Model
Term mapped from: InterPro:cd03464
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0008199 | ferric iron binding |
Inferred from Sequence Model
Term mapped from: InterPro:PF00775
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0003824 | catalytic activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF00775
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen |
Inferred from Sequence Model
Term mapped from: InterPro:SSF49482
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0055114 | oxidation-reduction process |
Inferred from Sequence Model
Term mapped from: InterPro:cd03464
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006725 | cellular aromatic compound metabolic process |
Inferred from Sequence Model
Term mapped from: InterPro:PF00775
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0005506 | iron ion binding |
Inferred from Sequence Model
Term mapped from: InterPro:cd03464
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | bcj01220 | Degradation of aromatic compounds | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | bcj00362 | Benzoate degradation | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG (InterPro) | 00624 | Polycyclic aromatic hydrocarbon degradation | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
|
| KEGG | bcj01120 | Microbial metabolism in diverse environments | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | bcj00624 | Polycyclic aromatic hydrocarbon degradation | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | bcj01100 | Metabolic pathways | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG (InterPro) | 00362 | Benzoate degradation | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
|
| MetaCyc | 4-sulfocatechol degradation | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Gene3D | G3DSA:2.60.130.10 | IPR015889 | Intradiol ring-cleavage dioxygenase, core | 2 | 235 | 2.0E-83 | |
| SUPERFAMILY | SSF49482 | IPR015889 | Intradiol ring-cleavage dioxygenase, core | 6 | 234 | 1.11E-71 | |
| CDD | cd03464 | 3,4-PCD_beta | IPR012785 | Protocatechuate 3,4-dioxygenase, beta subunit | 10 | 230 | 8.36555E-151 |
| Pfam | PF12391 | Protocatechuate 3,4-dioxygenase beta subunit N terminal | IPR024756 | Protocatechuate 3,4-dioxygenase beta subunit, N-terminal | 9 | 40 | 1.5E-13 |
| ProSitePatterns | PS00083 | Intradiol ring-cleavage dioxygenases signature. | IPR000627 | Intradiol ring-cleavage dioxygenase, C-terminal | 79 | 107 | - |
| Pfam | PF00775 | Dioxygenase | IPR000627 | Intradiol ring-cleavage dioxygenase, C-terminal | 47 | 227 | 9.1E-63 |
| TIGRFAM | TIGR02422 | protocat_beta: protocatechuate 3,4-dioxygenase, beta subunit | IPR012785 | Protocatechuate 3,4-dioxygenase, beta subunit | 15 | 234 | 4.4E-109 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.