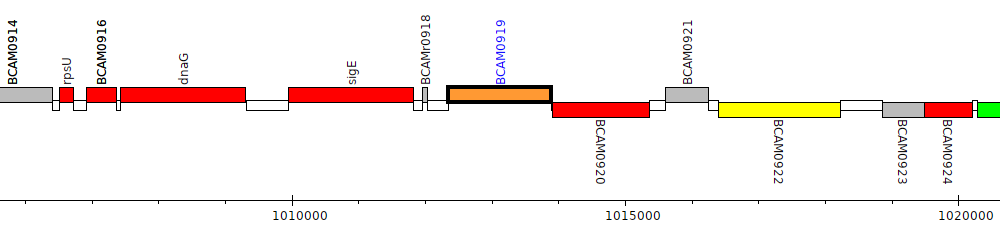
Burkholderia cenocepacia J2315, BCAM0919
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0003824 | catalytic activity |
Inferred from Sequence Model
Term mapped from: InterPro:SM00155
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Gene3D | G3DSA:3.30.870.10 | 56 | 238 | 1.4E-37 | |||
| Pfam | PF13091 | PLD-like domain | IPR025202 | Phospholipase D-like domain | 77 | 222 | 4.2E-16 |
| SUPERFAMILY | SSF56024 | 283 | 460 | 3.81E-33 | |||
| Gene3D | G3DSA:3.30.870.10 | 294 | 473 | 4.8E-37 | |||
| CDD | cd09111 | PLDc_ymdC_like_1 | 62 | 223 | 1.78347E-87 | ||
| Pfam | PF13091 | PLD-like domain | IPR025202 | Phospholipase D-like domain | 317 | 453 | 3.9E-24 |
| ProSiteProfiles | PS51257 | Prokaryotic membrane lipoprotein lipid attachment site profile. | 1 | 19 | 5.0 | ||
| SMART | SM00155 | IPR001736 | Phospholipase D/Transphosphatidylase | 402 | 429 | 5.2E-4 | |
| CDD | cd09113 | PLDc_ymdC_like_2 | 289 | 511 | 3.82409E-96 | ||
| ProSiteProfiles | PS50035 | Phospholipase D phosphodiesterase active site profile. | IPR001736 | Phospholipase D/Transphosphatidylase | 163 | 190 | 12.543 |
| SUPERFAMILY | SSF56024 | 37 | 234 | 6.95E-47 | |||
| SMART | SM00155 | IPR001736 | Phospholipase D/Transphosphatidylase | 163 | 190 | 9.0E-5 | |
| ProSiteProfiles | PS50035 | Phospholipase D phosphodiesterase active site profile. | IPR001736 | Phospholipase D/Transphosphatidylase | 402 | 429 | 12.985 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.