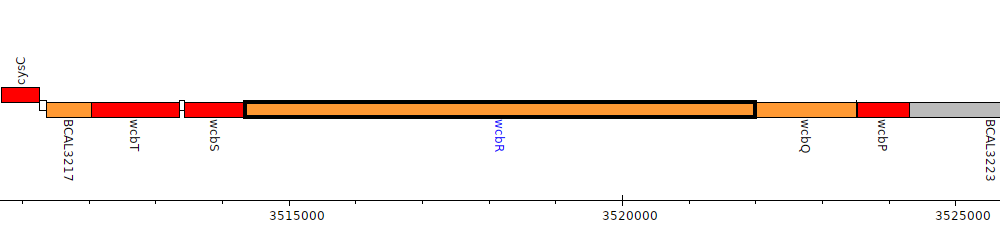
Burkholderia cenocepacia J2315, BCAL3220 (wcbR)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0055114 | oxidation-reduction process |
Inferred from Sequence Model
Term mapped from: InterPro:PF00107
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0003824 | catalytic activity |
Inferred from Sequence Model
Term mapped from: InterPro:G3DSA:3.40.47.10
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016491 | oxidoreductase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PS01162
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0008270 | zinc ion binding |
Inferred from Sequence Model
Term mapped from: InterPro:PS01162
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0031177 | phosphopantetheine binding |
Inferred from Sequence Model
Term mapped from: InterPro:SM00823
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016740 | transferase activity |
Inferred from Sequence Model
Term mapped from: InterPro:G3DSA:3.40.366.10
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Pfam | PF00698 | Acyl transferase domain | IPR014043 | Acyl transferase | 541 | 857 | 2.6E-62 |
| SUPERFAMILY | SSF55048 | IPR016036 | Malonyl-CoA ACP transacylase, ACP-binding | 672 | 738 | 1.96E-10 | |
| SUPERFAMILY | SSF53901 | IPR016039 | Thiolase-like | 2 | 427 | 6.24E-100 | |
| Pfam | PF00109 | Beta-ketoacyl synthase, N-terminal domain | IPR014030 | Beta-ketoacyl synthase, N-terminal | 3 | 252 | 6.1E-90 |
| Gene3D | G3DSA:3.90.180.10 | 1790 | 2132 | 4.3E-118 | |||
| Pfam | PF16197 | Ketoacyl-synthetase C-terminal extension | IPR032821 | Ketoacyl-synthetase, C-terminal extension | 381 | 497 | 9.4E-19 |
| Pfam | PF00107 | Zinc-binding dehydrogenase | IPR013149 | Alcohol dehydrogenase, C-terminal | 1942 | 2054 | 1.5E-23 |
| SMART | SM00825 | Beta-ketoacyl synthase | IPR020841 | Polyketide synthase, beta-ketoacyl synthase domain | 5 | 431 | 3.9E-214 |
| Pfam | PF13489 | Methyltransferase domain | 1383 | 1548 | 1.0E-8 | ||
| Pfam | PF14765 | Polyketide synthase dehydratase | IPR020807 | Polyketide synthase, dehydratase domain | 910 | 1190 | 5.3E-54 |
| Gene3D | G3DSA:3.40.366.10 | IPR001227 | Acyl transferase domain superfamily | 539 | 851 | 4.1E-122 | |
| CDD | cd00833 | PKS | 5 | 426 | 0.0 | ||
| SMART | SM00823 | Phosphopantetheine attachment site | IPR020806 | Polyketide synthase, phosphopantetheine-binding domain | 2419 | 2490 | 1.3E-8 |
| Gene3D | G3DSA:3.30.70.3290 | 455 | 894 | 4.1E-122 | |||
| SUPERFAMILY | SSF53335 | IPR029063 | S-adenosyl-L-methionine-dependent methyltransferase | 1383 | 1553 | 1.1E-13 | |
| SMART | SM00826 | IPR020807 | Polyketide synthase, dehydratase domain | 910 | 1072 | 2.5E-39 | |
| Gene3D | G3DSA:3.40.50.720 | 1575 | 1789 | 4.4E-28 | |||
| Gene3D | G3DSA:3.10.129.110 | IPR042104 | Polyketide synthase, dehydratase domain superfamily | 901 | 1193 | 1.9E-55 | |
| Gene3D | G3DSA:3.40.47.10 | IPR016039 | Thiolase-like | 1 | 454 | 1.1E-187 | |
| SUPERFAMILY | SSF52151 | IPR016035 | Acyl transferase/acyl hydrolase/lysophospholipase | 537 | 839 | 1.23E-61 | |
| SUPERFAMILY | SSF51735 | IPR036291 | NAD(P)-binding domain superfamily | 1589 | 1774 | 4.01E-29 | |
| SMART | SM00827 | Acyl transferase domain in polyketide synthase (PKS) enzymes. | IPR020801 | Polyketide synthase, acyl transferase domain | 541 | 848 | 5.8E-94 |
| SMART | SM00822 | This enzymatic domain is part of bacterial polyketide synthases and catalyses the first step in the reductive modification of the beta-carbonyl centres in the growing polyketide chain. It uses NADPH to reduce the keto group to a hydroxy group. | 2138 | 2319 | 2.6E-48 | ||
| Gene3D | G3DSA:3.40.50.150 | 1353 | 1559 | 9.6E-14 | |||
| Gene3D | G3DSA:1.10.1200.10 | IPR036736 | ACP-like superfamily | 2404 | 2496 | 2.3E-11 | |
| ProSiteProfiles | PS50075 | Carrier protein (CP) domain profile. | IPR009081 | Phosphopantetheine binding ACP domain | 2413 | 2490 | 12.807 |
| Gene3D | G3DSA:3.40.50.720 | 2133 | 2390 | 5.1E-59 | |||
| SMART | SM01294 | 2420 | 2486 | 0.0017 | |||
| ProSitePatterns | PS00606 | Beta-ketoacyl synthases active site. | IPR018201 | Beta-ketoacyl synthase, active site | 165 | 181 | - |
| Pfam | PF00550 | Phosphopantetheine attachment site | IPR009081 | Phosphopantetheine binding ACP domain | 2421 | 2486 | 9.6E-11 |
| SUPERFAMILY | SSF47336 | IPR036736 | ACP-like superfamily | 2418 | 2507 | 3.66E-13 | |
| Pfam | PF08659 | KR domain | IPR013968 | Polyketide synthase, ketoreductase domain | 2138 | 2318 | 3.0E-60 |
| Pfam | PF02801 | Beta-ketoacyl synthase, C-terminal domain | IPR014031 | Beta-ketoacyl synthase, C-terminal | 260 | 378 | 2.2E-35 |
| ProSitePatterns | PS01162 | Quinone oxidoreductase / zeta-crystallin signature. | IPR002364 | Quinone oxidoreductase/zeta-crystallin, conserved site | 1931 | 1952 | - |
| SMART | SM00829 | IPR020843 | Polyketide synthase, enoylreductase domain | 1798 | 2113 | 3.2E-176 | |
| SUPERFAMILY | SSF50129 | IPR011032 | GroES-like superfamily | 1795 | 1936 | 9.75E-28 | |
| SUPERFAMILY | SSF51735 | IPR036291 | NAD(P)-binding domain superfamily | 2138 | 2341 | 2.98E-37 | |
| CDD | cd05195 | enoyl_red | 1816 | 2113 | 3.32209E-120 | ||
| SUPERFAMILY | SSF51735 | IPR036291 | NAD(P)-binding domain superfamily | 1903 | 2057 | 1.42E-48 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.