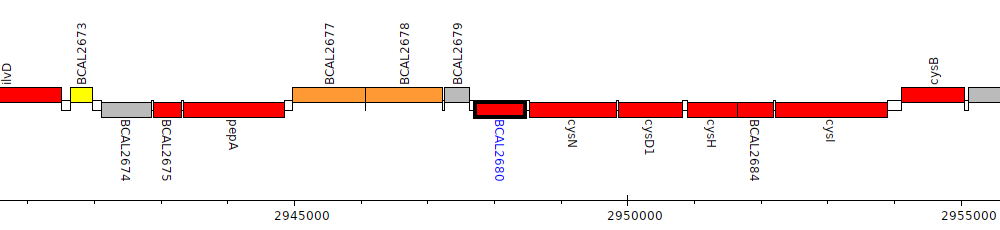
Burkholderia cenocepacia J2315, BCAL2680
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0019354 | siroheme biosynthetic process |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR01469
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0008168 | methyltransferase activity |
Inferred from Sequence Model
Term mapped from: InterPro:G3DSA:3.40.1010.10
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0055114 | oxidation-reduction process |
Inferred from Sequence Model
Term mapped from: InterPro:PS00839
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | bcj00860 | Porphyrin and chlorophyll metabolism | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| MetaCyc | siroheme biosynthesis | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
||
| KEGG (InterPro) | 00860 | Porphyrin and chlorophyll metabolism | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
|
| KEGG | bcj01100 | Metabolic pathways | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| MetaCyc | cob(II)yrinate <i>a,c</i>-diamide biosynthesis I (early cobalt insertion) | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
||
| MetaCyc | factor 430 biosynthesis | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
||
| KEGG | bcj01110 | Biosynthesis of secondary metabolites | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| CDD | cd11642 | SUMT | IPR006366 | Uroporphyrin-III C-methyltransferase | 3 | 231 | 3.04154E-91 |
| TIGRFAM | TIGR01469 | cobA_cysG_Cterm: uroporphyrinogen-III C-methyltransferase | IPR006366 | Uroporphyrin-III C-methyltransferase | 2 | 233 | 3.2E-80 |
| ProSitePatterns | PS00839 | Uroporphyrin-III C-methyltransferase signature 1. | IPR003043 | Uroporphiryn-III C-methyltransferase, conserved site | 7 | 21 | - |
| Gene3D | G3DSA:3.40.1010.10 | IPR014777 | Tetrapyrrole methylase, subdomain 1 | 1 | 116 | 7.3E-42 | |
| ProSitePatterns | PS00840 | Uroporphyrin-III C-methyltransferase signature 2. | IPR003043 | Uroporphiryn-III C-methyltransferase, conserved site | 81 | 114 | - |
| Gene3D | G3DSA:3.30.950.10 | IPR014776 | Tetrapyrrole methylase, subdomain 2 | 117 | 247 | 9.9E-26 | |
| Pfam | PF00590 | Tetrapyrrole (Corrin/Porphyrin) Methylases | IPR000878 | Tetrapyrrole methylase | 3 | 209 | 4.9E-46 |
| SUPERFAMILY | SSF53790 | IPR035996 | Tetrapyrrole methylase superfamily | 1 | 237 | 6.8E-62 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.