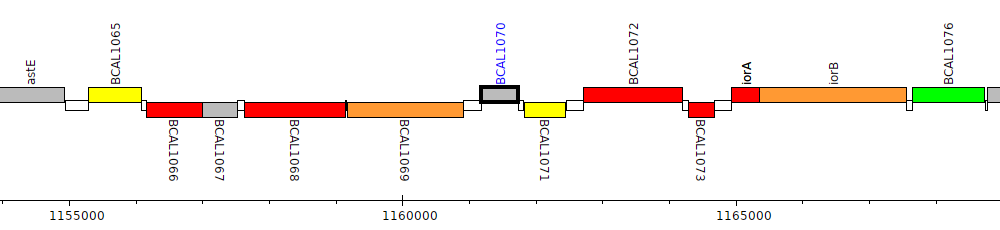
Burkholderia cenocepacia J2315, BCAL1070
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0016209 | antioxidant activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF00578
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0045454 | cell redox homeostasis |
Inferred from Sequence Model
Term mapped from: InterPro:PS51352
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0055114 | oxidation-reduction process |
Inferred from Sequence Model
Term mapped from: InterPro:PF00578
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016491 | oxidoreductase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF00578
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG (InterPro) | 00480 | Glutathione metabolism | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Pfam | PF00578 | AhpC/TSA family | IPR000866 | Alkyl hydroperoxide reductase subunit C/ Thiol specific antioxidant | 28 | 154 | 6.1E-27 |
| SUPERFAMILY | SSF52833 | IPR036249 | Thioredoxin-like superfamily | 25 | 175 | 4.12E-40 | |
| ProSiteProfiles | PS51352 | Thioredoxin domain profile. | IPR013766 | Thioredoxin domain | 25 | 177 | 14.832 |
| CDD | cd03017 | PRX_BCP | 29 | 170 | 2.39337E-50 | ||
| PIRSF | PIRSF000239 | IPR024706 | Peroxiredoxin, AhpC-type | 21 | 179 | 7.2E-21 | |
| Gene3D | G3DSA:3.40.30.10 | 19 | 177 | 3.8E-36 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.