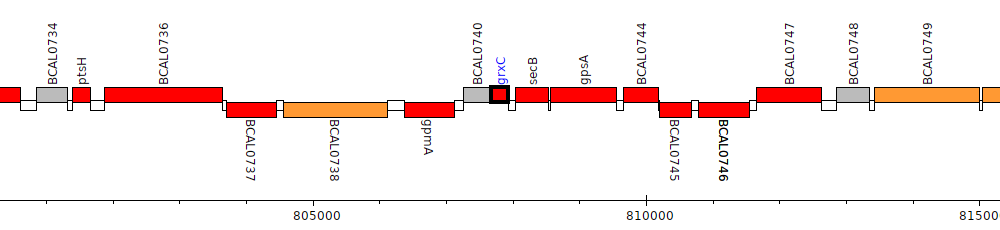
Burkholderia cenocepacia J2315, BCAL0741 (grxC)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0009055 | electron transfer activity |
Inferred from Sequence Model
Term mapped from: InterPro:PS00195
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0045454 | cell redox homeostasis |
Inferred from Sequence Model
Term mapped from: InterPro:PS00195
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0015035 | protein disulfide oxidoreductase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF00462
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Gene3D | G3DSA:3.40.30.10 | 1 | 86 | 3.2E-25 | |||
| PRINTS | PR00160 | Glutaredoxin signature | IPR014025 | Glutaredoxin subgroup | 61 | 74 | 9.0E-15 |
| Pfam | PF00462 | Glutaredoxin | IPR002109 | Glutaredoxin | 4 | 64 | 3.5E-18 |
| ProSiteProfiles | PS51354 | Glutaredoxin domain profile. | IPR002109 | Glutaredoxin | 1 | 86 | 20.679 |
| ProSitePatterns | PS00195 | Glutaredoxin active site. | IPR011767 | Glutaredoxin active site | 6 | 22 | - |
| SUPERFAMILY | SSF52833 | IPR036249 | Thioredoxin-like superfamily | 1 | 84 | 1.35E-25 | |
| CDD | cd03418 | GRX_GRXb_1_3_like | IPR011900 | Glutaredoxin, GrxC | 3 | 77 | 1.78051E-34 |
| TIGRFAM | TIGR02181 | GRX_bact: glutaredoxin 3 | IPR011900 | Glutaredoxin, GrxC | 4 | 83 | 8.5E-32 |
| PRINTS | PR00160 | Glutaredoxin signature | IPR014025 | Glutaredoxin subgroup | 47 | 60 | 9.0E-15 |
| PRINTS | PR00160 | Glutaredoxin signature | IPR014025 | Glutaredoxin subgroup | 4 | 22 | 9.0E-15 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.