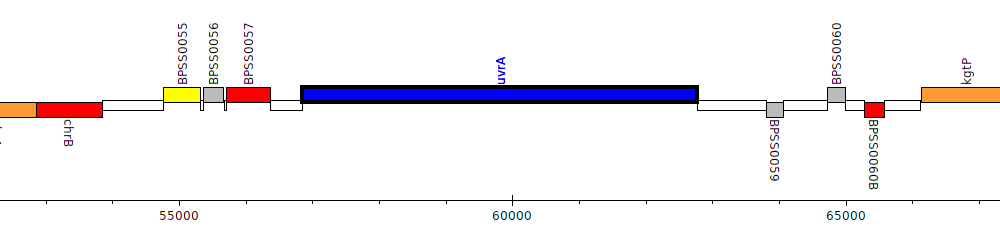
Burkholderia pseudomallei K96243, BPSS0058 (uvrA)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Cellular Component | GO:0009380 | excinuclease repair complex |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR00630
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0005524 | ATP binding |
Inferred from Sequence Model
Term mapped from: InterPro:PS50893
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0005524 | ATP binding |
Inferred from Sequence Model
Term mapped from: InterPro:PS50893
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016887 | ATPase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PS50893
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006289 | nucleotide-excision repair |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR00630
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006289 | nucleotide-excision repair |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR00630
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0003677 | DNA binding |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR00630
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0003677 | DNA binding |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR00630
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016887 | ATPase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PS50893
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Cellular Component | GO:0009380 | excinuclease repair complex |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR00630
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | bps03420 | Nucleotide excision repair | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Pfam | PF17755 | UvrA DNA-binding domain | IPR041552 | UvrA DNA-binding domain | 297 | 361 | 7.0E-8 |
| Gene3D | G3DSA:3.40.50.300 | 646 | 984 | 3.9E-125 | |||
| SMART | SM00382 | IPR003593 | AAA+ ATPase domain | 1596 | 1918 | 2.8 | |
| Gene3D | G3DSA:1.20.1580.10 | 721 | 863 | 3.9E-125 | |||
| Gene3D | G3DSA:3.40.50.300 | 1298 | 1552 | 5.6E-56 | |||
| Gene3D | G3DSA:1.20.1580.10 | 1717 | 1883 | 2.5E-130 | |||
| Gene3D | G3DSA:3.30.190.20 | 1147 | 1266 | 2.4E-28 | |||
| Gene3D | G3DSA:3.30.190.20 | 133 | 237 | 6.3E-202 | |||
| ProSitePatterns | PS00211 | ABC transporters family signature. | IPR017871 | ABC transporter, conserved site | 1833 | 1847 | - |
| SUPERFAMILY | SSF52540 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 1578 | 1926 | 3.53E-42 | |
| TIGRFAM | TIGR00630 | uvra: excinuclease ABC subunit A | IPR004602 | UvrABC system subunit A | 7 | 972 | 1.9E-269 |
| ProSitePatterns | PS00211 | ABC transporters family signature. | IPR017871 | ABC transporter, conserved site | 863 | 877 | - |
| SUPERFAMILY | SSF52540 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 6 | 597 | 7.72E-39 | |
| Gene3D | G3DSA:3.40.50.300 | 1012 | 1133 | 1.6E-21 | |||
| Gene3D | G3DSA:3.40.50.300 | 9 | 622 | 6.3E-202 | |||
| Gene3D | G3DSA:1.20.1580.10 | 93 | 520 | 6.3E-202 | |||
| MobiDBLite | mobidb-lite | consensus disorder prediction | 1334 | 1349 | - | ||
| SMART | SM00382 | IPR003593 | AAA+ ATPase domain | 1045 | 1519 | 4.8 | |
| SMART | SM00382 | IPR003593 | AAA+ ATPase domain | 662 | 954 | 0.96 | |
| MobiDBLite | mobidb-lite | consensus disorder prediction | 1334 | 1364 | - | ||
| Gene3D | G3DSA:3.40.50.300 | 1580 | 1948 | 2.5E-130 | |||
| Gene3D | G3DSA:1.10.8.280 | 294 | 399 | 6.3E-202 | |||
| Pfam | PF17760 | UvrA interaction domain | IPR041102 | UvrA, interaction domain | 1152 | 1255 | 4.2E-19 |
| ProSitePatterns | PS00211 | ABC transporters family signature. | IPR017871 | ABC transporter, conserved site | 1442 | 1456 | - |
| CDD | cd03270 | ABC_UvrA_I | 1418 | 1530 | 3.06657E-62 | ||
| ProSiteProfiles | PS50893 | ATP-binding cassette, ABC transporter-type domain profile. | IPR003439 | ABC transporter-like | 1020 | 1542 | 8.83 |
| ProSiteProfiles | PS50893 | ATP-binding cassette, ABC transporter-type domain profile. | IPR003439 | ABC transporter-like | 195 | 615 | 9.331 |
| SUPERFAMILY | SSF52540 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 648 | 957 | 4.14E-36 | |
| SUPERFAMILY | SSF52540 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 1028 | 1524 | 1.22E-39 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.