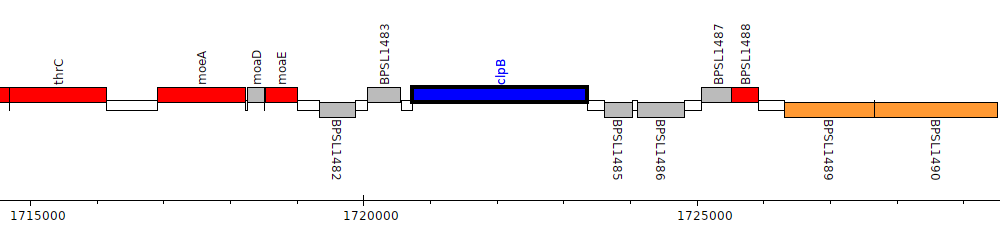
Burkholderia pseudomallei K96243, BPSL1484 (clpB)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0019538 | protein metabolic process |
Inferred from Sequence Model
Term mapped from: InterPro:SSF81923
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0042026 | protein refolding |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR03346
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Cellular Component | GO:0005737 | cytoplasm |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR03346
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0005524 | ATP binding |
Inferred from Sequence Model
Term mapped from: InterPro:PS00870
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0009408 | response to heat |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR03346
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| PRINTS | PR00300 | ATP-dependent Clp protease ATP-binding subunit signature | IPR001270 | ClpA/B family | 692 | 710 | 1.3E-42 |
| Gene3D | G3DSA:3.40.50.300 | 566 | 780 | 4.9E-89 | |||
| Pfam | PF17871 | AAA lid domain | IPR041546 | ClpA/ClpB, AAA lid domain | 353 | 453 | 5.4E-36 |
| PRINTS | PR00300 | ATP-dependent Clp protease ATP-binding subunit signature | IPR001270 | ClpA/B family | 725 | 739 | 1.3E-42 |
| PRINTS | PR00300 | ATP-dependent Clp protease ATP-binding subunit signature | IPR001270 | ClpA/B family | 663 | 681 | 1.3E-42 |
| SUPERFAMILY | SSF81923 | IPR036628 | Clp, N-terminal domain superfamily | 15 | 167 | 8.83E-41 | |
| CDD | cd00009 | AAA | 192 | 352 | 8.42898E-23 | ||
| Pfam | PF02861 | Clp amino terminal domain, pathogenicity island component | IPR004176 | Clp, N-terminal | 28 | 79 | 1.4E-12 |
| Gene3D | G3DSA:1.10.1780.10 | IPR036628 | Clp, N-terminal domain superfamily | 12 | 157 | 2.9E-34 | |
| Pfam | PF02861 | Clp amino terminal domain, pathogenicity island component | IPR004176 | Clp, N-terminal | 105 | 156 | 6.0E-9 |
| Gene3D | G3DSA:3.40.50.300 | 354 | 561 | 8.3E-68 | |||
| Pfam | PF07724 | AAA domain (Cdc48 subfamily) | IPR003959 | ATPase, AAA-type, core | 613 | 775 | 7.6E-55 |
| Pfam | PF10431 | C-terminal, D2-small domain, of ClpB protein | IPR019489 | Clp ATPase, C-terminal | 782 | 860 | 2.3E-22 |
| Coils | Coil | 445 | 472 | - | |||
| SMART | SM01086 | IPR019489 | Clp ATPase, C-terminal | 782 | 871 | 1.3E-32 | |
| ProSitePatterns | PS00870 | Chaperonins clpA/B signature 1. | IPR018368 | ClpA/B, conserved site 1 | 305 | 317 | - |
| Pfam | PF00004 | ATPase family associated with various cellular activities (AAA) | IPR003959 | ATPase, AAA-type, core | 214 | 346 | 4.1E-14 |
| Coils | Coil | 423 | 443 | - | |||
| SUPERFAMILY | SSF52540 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 170 | 561 | 2.22E-99 | |
| SMART | SM00382 | IPR003593 | AAA+ ATPase domain | 209 | 354 | 1.8E-12 | |
| Gene3D | G3DSA:1.10.8.60 | 782 | 875 | 3.5E-25 | |||
| Coils | Coil | 483 | 503 | - | |||
| PRINTS | PR00300 | ATP-dependent Clp protease ATP-binding subunit signature | IPR001270 | ClpA/B family | 618 | 636 | 1.3E-42 |
| SUPERFAMILY | SSF52540 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 563 | 870 | 7.47E-82 | |
| Gene3D | G3DSA:3.40.50.300 | 158 | 353 | 5.5E-76 | |||
| ProSitePatterns | PS00871 | Chaperonins clpA/B signature 2. | IPR028299 | ClpA/B, conserved site 2 | 648 | 666 | - |
| CDD | cd00009 | AAA | 588 | 779 | 5.29129E-21 | ||
| TIGRFAM | TIGR03346 | chaperone_ClpB: ATP-dependent chaperone protein ClpB | IPR017730 | Chaperonin ClpB | 17 | 871 | 0.0 |
| SMART | SM00382 | IPR003593 | AAA+ ATPase domain | 614 | 784 | 5.5E-11 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.