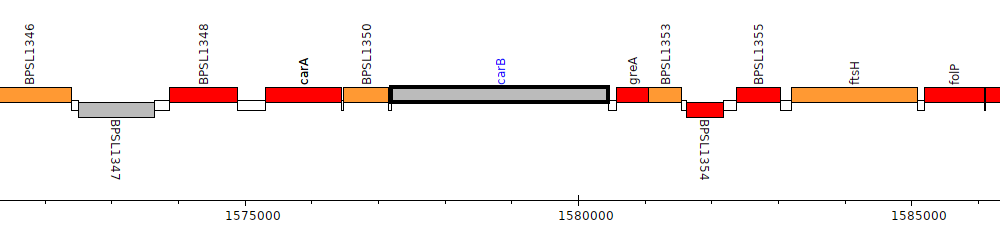
Burkholderia pseudomallei K96243, BPSL1351 (carB)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0046872 | metal ion binding |
Inferred from Sequence Model
Term mapped from: InterPro:PS50975
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0005524 | ATP binding |
Inferred from Sequence Model
Term mapped from: InterPro:PS00866
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006807 | nitrogen compound metabolic process |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR01369
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| MetaCyc | L-arginine biosynthesis IV (archaebacteria) | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
||
| KEGG | bps00240 | Pyrimidine metabolism | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| MetaCyc | UMP biosynthesis I | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
||
| KEGG | bps00250 | Alanine, aspartate and glutamate metabolism | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| MetaCyc | UMP biosynthesis III | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
||
| KEGG (InterPro) | 00250 | Alanine, aspartate and glutamate metabolism | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
|
| KEGG (InterPro) | 00240 | Pyrimidine metabolism | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
|
| MetaCyc | L-arginine biosynthesis III (via <i>N</i>-acetyl-L-citrulline) | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
||
| KEGG | bps01100 | Metabolic pathways | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| MetaCyc | UMP biosynthesis II | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| PRINTS | PR00098 | Carbamoyl-phosphate synthase protein CPSase domain signature | IPR005483 | Carbamoyl-phosphate synthase large subunit, CPSase domain | 387 | 405 | 2.1E-66 |
| Gene3D | G3DSA:3.40.50.20 | 1 | 116 | 3.7E-50 | |||
| PRINTS | PR00098 | Carbamoyl-phosphate synthase protein CPSase domain signature | IPR005483 | Carbamoyl-phosphate synthase large subunit, CPSase domain | 211 | 230 | 2.1E-66 |
| ProSitePatterns | PS00866 | Carbamoyl-phosphate synthase subdomain signature 1. | IPR005479 | Carbamoyl-phosphate synthetase large subunit-like, ATP-binding domain | 171 | 185 | - |
| SUPERFAMILY | SSF52440 | IPR016185 | Pre-ATP-grasp domain superfamily | 1 | 127 | 2.59E-51 | |
| Pfam | PF02142 | MGS-like domain | IPR011607 | Methylglyoxal synthase-like domain | 967 | 1053 | 1.0E-20 |
| Hamap | MF_01210_B | Carbamoyl-phosphate synthase large chain [carB]. | IPR006275 | Carbamoyl-phosphate synthase, large subunit | 1 | 1074 | 13.171 |
| SUPERFAMILY | SSF52440 | IPR016185 | Pre-ATP-grasp domain superfamily | 559 | 677 | 9.93E-50 | |
| Coils | Coil | 1076 | 1084 | - | |||
| SMART | SM01096 | IPR005480 | Carbamoyl-phosphate synthetase, large subunit oligomerisation domain | 426 | 549 | 1.3E-74 | |
| Hamap | MF_01210_A | Carbamoyl-phosphate synthase large chain [carB]. | IPR006275 | Carbamoyl-phosphate synthase, large subunit | 5 | 1079 | 13.354 |
| PRINTS | PR00098 | Carbamoyl-phosphate synthase protein CPSase domain signature | IPR005483 | Carbamoyl-phosphate synthase large subunit, CPSase domain | 175 | 187 | 2.1E-66 |
| ProSitePatterns | PS00866 | Carbamoyl-phosphate synthase subdomain signature 1. | IPR005479 | Carbamoyl-phosphate synthetase large subunit-like, ATP-binding domain | 711 | 725 | - |
| ProSitePatterns | PS00867 | Carbamoyl-phosphate synthase subdomain signature 2. | IPR005479 | Carbamoyl-phosphate synthetase large subunit-like, ATP-binding domain | 849 | 856 | - |
| PRINTS | PR00098 | Carbamoyl-phosphate synthase protein CPSase domain signature | IPR005483 | Carbamoyl-phosphate synthase large subunit, CPSase domain | 246 | 263 | 2.1E-66 |
| SMART | SM00851 | IPR011607 | Methylglyoxal synthase-like domain | 966 | 1053 | 1.2E-30 | |
| PRINTS | PR00098 | Carbamoyl-phosphate synthase protein CPSase domain signature | IPR005483 | Carbamoyl-phosphate synthase large subunit, CPSase domain | 48 | 58 | 2.1E-66 |
| ProSitePatterns | PS00867 | Carbamoyl-phosphate synthase subdomain signature 2. | IPR005479 | Carbamoyl-phosphate synthetase large subunit-like, ATP-binding domain | 304 | 311 | - |
| Gene3D | G3DSA:3.40.50.1380 | IPR036914 | Methylglyoxal synthase-like domain superfamily | 947 | 1084 | 1.2E-36 | |
| ProSiteProfiles | PS50975 | ATP-grasp fold profile. | IPR011761 | ATP-grasp fold | 133 | 335 | 42.857 |
| ProSiteProfiles | PS51257 | Prokaryotic membrane lipoprotein lipid attachment site profile. | 1 | 24 | 6.0 | ||
| Gene3D | G3DSA:3.40.50.20 | 556 | 664 | 1.4E-48 | |||
| Gene3D | G3DSA:3.30.470.20 | 119 | 409 | 8.0E-129 | |||
| TIGRFAM | TIGR01369 | CPSaseII_lrg: carbamoyl-phosphate synthase, large subunit | IPR006275 | Carbamoyl-phosphate synthase, large subunit | 2 | 1063 | 0.0 |
| SUPERFAMILY | SSF56059 | 675 | 944 | 1.29E-100 | |||
| Pfam | PF02786 | Carbamoyl-phosphate synthase L chain, ATP binding domain | IPR005479 | Carbamoyl-phosphate synthetase large subunit-like, ATP-binding domain | 128 | 341 | 1.3E-70 |
| SUPERFAMILY | SSF48108 | IPR036897 | Carbamoyl-phosphate synthetase, large subunit oligomerisation domain superfamily | 403 | 556 | 7.85E-53 | |
| Pfam | PF02786 | Carbamoyl-phosphate synthase L chain, ATP binding domain | IPR005479 | Carbamoyl-phosphate synthetase large subunit-like, ATP-binding domain | 675 | 886 | 3.4E-32 |
| ProSiteProfiles | PS50975 | ATP-grasp fold profile. | IPR011761 | ATP-grasp fold | 680 | 880 | 45.729 |
| Gene3D | G3DSA:1.10.1030.10 | IPR036897 | Carbamoyl-phosphate synthetase, large subunit oligomerisation domain superfamily | 410 | 555 | 1.0E-46 | |
| ProSiteProfiles | PS51855 | MGS-like domain profile. | 947 | 1084 | 28.283 | ||
| Gene3D | G3DSA:3.30.470.20 | 667 | 946 | 3.0E-115 | |||
| PRINTS | PR00098 | Carbamoyl-phosphate synthase protein CPSase domain signature | IPR005483 | Carbamoyl-phosphate synthase large subunit, CPSase domain | 304 | 333 | 2.1E-66 |
| PRINTS | PR00098 | Carbamoyl-phosphate synthase protein CPSase domain signature | IPR005483 | Carbamoyl-phosphate synthase large subunit, CPSase domain | 19 | 33 | 2.1E-66 |
| CDD | cd01424 | MGS_CPS_II | IPR033937 | Carbamoyl-phosphate synthase large chain, methylglyoxal synthase-like domain | 953 | 1063 | 4.09595E-39 |
| Pfam | PF02787 | Carbamoyl-phosphate synthetase large chain, oligomerisation domain | IPR005480 | Carbamoyl-phosphate synthetase, large subunit oligomerisation domain | 429 | 549 | 1.1E-38 |
| SUPERFAMILY | SSF56059 | 128 | 408 | 2.83E-107 | |||
| SUPERFAMILY | SSF52335 | IPR036914 | Methylglyoxal synthase-like domain superfamily | 946 | 1082 | 3.92E-38 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.