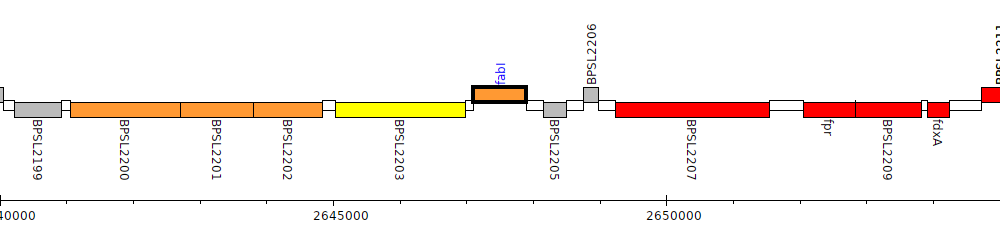
Burkholderia pseudomallei K96243, BPSL2204 (fabI)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Feature Overview
| Strain |
Burkholderia pseudomallei K96243 - Assembly GCF_000011545.1
GCF_000011545.1|latest |
| Locus Tag |
BPSL2204
|
| Name |
fabI
|
| Replicon | chromosome 1 |
| Genomic location | 2647102 - 2647893 (+ strand) |
Cross-References
| RefSeq | YP_108799.1 |
| GI | 53719813 |
| Entrez | 3091461 |
| GI | 53719813 |
| INSDC | CAH36206.1 |
| NCBI Locus Tag | BPSL2204 |
| RefSeq | YP_108799.1,GeneID |
| UniParc | UPI000043B2BE |
| UniProtKB Acc | Q63SW7 |
| UniProtKB ID | Q63SW7_BURPS |
| UniRef100 | UniRef100_A0A069B9A4 |
| UniRef50 | UniRef50_P16657 |
| UniRef90 | UniRef90_N0AC56 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
enoyl-ACP reductase
|
| Synonyms | |
| Evidence for Translation | |
| Charge (pH 7) | -6.10 |
| Kyte-Doolittle Hydrophobicity Value | 0.247 |
| Molecular Weight (kDa) | 27770.3 |
| Isoelectric Point (pI) | 4.96 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
PDB 3D Structures
| Accession | Header | Accession Date | Compound | Source | Resolution | Method | Percent Identity |
| 4BKU | OXIDOREDUCTASE | 04/29/13 | Enoyl-ACP reductase FabI from Burkholderia pseudomallei with cofactor NADH and inhibitor PT155 | BURKHOLDERIA PSEUDOMALLEI | 1.841 | X-RAY DIFFRACTION | 100.0 |
| 5I8Z | OXIDOREDUCTASE | 02/19/16 | Crystal structure of B. pseudomallei FabI in complex with NAD and PT12 | BURKHOLDERIA PSEUDOMALLEI | 1.623 | X-RAY DIFFRACTION | 100.0 |
| 5I9M | OXIDOREDUCTASE | 02/20/16 | Crystal structure of B. pseudomallei FabI in complex with NAD and PT408 | BURKHOLDERIA PSEUDOMALLEI | 2.25 | X-RAY DIFFRACTION | 100.0 |
| 5IFL | OXIDOREDUCTASE | 02/26/16 | Crystal structure of B. pseudomallei FabI in complex with NAD and triclosan | BURKHOLDERIA PSEUDOMALLEI | 2.6 | X-RAY DIFFRACTION | 100.0 |
| 5TRT | OXIDOREDUCTASE | 10/27/16 | Crystal Structure of enoyl-(acyl carrier protein) reductase from Burkholderia pseudomallei 1710b bound to NAD | BURKHOLDERIA PSEUDOMALLEI (STRAIN 1710B) | 1.85 | X-RAY DIFFRACTION | 100.0 |
| 5I9L | OXIDOREDUCTASE | 02/20/16 | Crystal structure of B. pseudomallei FabI in complex with NAD and PT404 | BURKHOLDERIA PSEUDOMALLEI | 1.8 | X-RAY DIFFRACTION | 100.0 |
| 5I7F | OXIDOREDUCTASE | 02/17/16 | Crystal structure of B. pseudomallei FabI in complex with NAD and PT405 | BURKHOLDERIA PSEUDOMALLEI | 2.7 | X-RAY DIFFRACTION | 100.0 |
| 3EK2 | OXIDOREDUCTASE | 09/18/08 | Crystal structure of eonyl-(acyl carrier protein) reductase from burkholderia pseudomallei 1719b | BURKHOLDERIA PSEUDOMALLEI 1710B | 1.9 | X-RAY DIFFRACTION | 100.0 |
| 4RLH | OXIDOREDUCTASE | 10/17/14 | Crystal structure of enoyl ACP reductase from Burkholderia pseudomallei in complex with AFN-1252 | BURKHOLDERIA PSEUDOMALLEI | 2.26 | X-RAY DIFFRACTION | 100.0 |
| 5I9N | OXIDOREDUCTASE | 02/20/16 | Crystal structure of B. pseudomallei FabI in complex with NAD and PT412 | BURKHOLDERIA PSEUDOMALLEI | 2.512 | X-RAY DIFFRACTION | 100.0 |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 605 genera
|
Orthologs/Comparative Genomics
| Burkholderia Ortholog Group |
BG019848 (328 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: BPSL2204
Search term: fabI
Search term: enoyl-ACP reductase
|
Human Homologs
References
|
Quantitative proteomic analysis of Burkholderia pseudomallei Bsa type III secretion system effectors using hypersecreting mutants.
Vander Broek CW, Chalmers KJ, Stevens MP, Stevens JM
Mol Cell Proteomics 2015 Apr;14(4):905-16
PubMed ID: 25635268
|