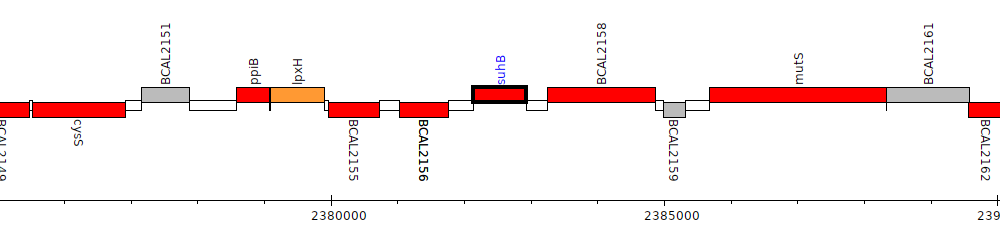
Burkholderia cenocepacia J2315, BCAL2157 (suhB)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0071978 | bacterial-type flagellum-dependent swarming motility |
Inferred from Sequence or Structural Similarity
Term mapped from: locus_tag:BURCENK562V_RS21000
|
ECO:0000250 sequence similarity evidence used in manual assertion |
22767545 | Reviewed by curator |
| Biological Process | GO:0033103 | protein secretion by the type VI secretion system |
Inferred from Sequence or Structural Similarity
Term mapped from: locus_tag:BURCENK562V_RS21000
|
ECO:0000250 sequence similarity evidence used in manual assertion |
22767545 | Reviewed by curator |
| Biological Process | GO:0044010 | single-species biofilm formation |
Inferred from Sequence or Structural Similarity
Term mapped from: locus_tag:BURCENK562V_RS21000
|
ECO:0000250 sequence similarity evidence used in manual assertion |
22767545 | Reviewed by curator |
| Biological Process | GO:0015628 | protein secretion by the type II secretion system |
Inferred from Sequence or Structural Similarity
Term mapped from: locus_tag:BURCENK562V_RS21000
|
ECO:0000250 sequence similarity evidence used in manual assertion |
22767545 | Reviewed by curator |
| Biological Process | GO:0046677 | response to antibiotic |
Inferred from Sequence or Structural Similarity
Term mapped from: locus_tag:BURCENK562V_RS21000
|
ECO:0000250 sequence similarity evidence used in manual assertion |
22767545 | Reviewed by curator |
| Biological Process | GO:0046854 | phosphatidylinositol phosphorylation |
Inferred from Sequence Model
Term mapped from: InterPro:PF00459
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0046855 | inositol phosphate dephosphorylation |
Inferred from Sequence Model
Term mapped from: InterPro:cd01639
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0008934 | inositol monophosphate 1-phosphatase activity |
Inferred from Sequence Model
Term mapped from: InterPro:cd01639
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | bcj01110 | Biosynthesis of secondary metabolites | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | bcj01100 | Metabolic pathways | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| MetaCyc | phytate degradation I | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
||
| KEGG | bcj00521 | Streptomycin biosynthesis | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG (InterPro) | 04070 | Phosphatidylinositol signaling system | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
|
| MetaCyc | D-<i>myo</i>-inositol (1,4,5)-trisphosphate degradation | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
||
| KEGG (InterPro) | 00562 | Inositol phosphate metabolism | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
|
| MetaCyc | <i>myo</i>-inositol biosynthesis | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
||
| KEGG (InterPro) | 00521 | Streptomycin biosynthesis | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
|
| KEGG | bcj00562 | Inositol phosphate metabolism | 73.0+/03-31, Mar 15 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| PRINTS | PR00377 | Inositol monophosphatase superfamily signature | IPR000760 | Inositol monophosphatase-like | 60 | 76 | 1.6E-61 |
| PRINTS | PR00377 | Inositol monophosphatase superfamily signature | IPR000760 | Inositol monophosphatase-like | 80 | 96 | 1.6E-61 |
| PRINTS | PR01959 | SuhB family inositol monophosphatase signature | IPR022337 | Inositol monophosphatase, SuhB | 17 | 39 | 1.4E-29 |
| ProSitePatterns | PS00629 | Inositol monophosphatase family signature 1. | IPR020583 | Inositol monophosphatase, metal-binding site | 80 | 93 | - |
| PRINTS | PR00377 | Inositol monophosphatase superfamily signature | IPR000760 | Inositol monophosphatase-like | 38 | 58 | 1.6E-61 |
| Gene3D | G3DSA:3.40.190.80 | 140 | 266 | 3.4E-35 | |||
| PRINTS | PR01959 | SuhB family inositol monophosphatase signature | IPR022337 | Inositol monophosphatase, SuhB | 103 | 122 | 1.4E-29 |
| Gene3D | G3DSA:3.30.540.10 | 1 | 139 | 7.4E-53 | |||
| PRINTS | PR00377 | Inositol monophosphatase superfamily signature | IPR000760 | Inositol monophosphatase-like | 207 | 231 | 1.6E-61 |
| ProSitePatterns | PS00630 | Inositol monophosphatase family signature 2. | IPR020550 | Inositol monophosphatase, conserved site | 210 | 224 | - |
| PRINTS | PR01959 | SuhB family inositol monophosphatase signature | IPR022337 | Inositol monophosphatase, SuhB | 155 | 181 | 1.4E-29 |
| SUPERFAMILY | SSF56655 | 1 | 260 | 2.36E-84 | |||
| CDD | cd01639 | IMPase | IPR033942 | Inositol monophosphatase | 4 | 245 | 2.68525E-116 |
| PRINTS | PR00377 | Inositol monophosphatase superfamily signature | IPR000760 | Inositol monophosphatase-like | 176 | 197 | 1.6E-61 |
| Pfam | PF00459 | Inositol monophosphatase family | IPR000760 | Inositol monophosphatase-like | 2 | 255 | 3.7E-66 |
| PRINTS | PR01959 | SuhB family inositol monophosphatase signature | IPR022337 | Inositol monophosphatase, SuhB | 197 | 209 | 1.4E-29 |
| PRINTS | PR00377 | Inositol monophosphatase superfamily signature | IPR000760 | Inositol monophosphatase-like | 129 | 152 | 1.6E-61 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.